library(gpttools)
crawl("https://r4ds.hadley.nz/")Large language models like GPT-3 and ChatGPT don’t need much of an introduction. They are enormously powerful with benchmarks being surpassed nearly as quickly as they are created. Despite the unprecedented model performance, these models struggle to provide accurate results when the appropriate response requires context more recent than the training data for a model. Vector databases can created from data sources outside the training corpus can address this gap by providing missing context to a model.
Vector databases can be use in semantic search with embeddings created form source text. By creating embeddings of text data and storing them in a database, we can quickly search for related documents and even perform advanced operations like similarity searches or clustering. This can be especially helpful when working with text data that is more context-dependent or domain-specific, such as scientific or technical documentation.
{gpttools} provides a set of tools for working with GPT-3 and other OpenAI models, including the ability to generate embeddings, perform similarity searches, and build vector databases. This package also has convenience functions to aid in scraping web pages to collect text data, generate embeddings, and store those embeddings in a vector database for future use.
To demonstrate the power of vector databases, we’ll use {gpttools} to build a vector database from R for Data Science. The approach uses semantic search to find the most relevant text from the book and then uses ChatGPT to generate a response based on that text via the recently release ChatGPT API.
Popular python packages such as llama-index and langchain provide easy utility functions to create vector stores for semantic search with a few lines of python code. {gpttools} aims to provide similar functionality with R using data frames as the data structure for the vector store.
Scraping Text from R4DS
The first step is to scrape the text from the R4DS book. We’ll use the crawl() function to scrape the text from the book and store it in a data frame. The crawl() function uses the rvest package to scrape the text from the online book and {tokenizers} to split the text into chunks for subsequent processing.
The code to scrape the data is relatively simple but is unlikely to work on all sites. From some internal testing, it works quite well on {pkgdown} and similar documentation sites.
Under the hood there are a few things going on. Here is the annotated function and associated functions:
crawl <- function(url,
index_create = TRUE,
aggressive = FALSE,
overwrite = FALSE,
num_cores = parallel::detectCores() - 1) {
local_domain <- urltools::url_parse(url)$domain
withr::local_options(list(
cli.progress_show_after = 0,
cli.progress_clear = FALSE
))
future::plan(future::multisession, workers = num_cores)
scraped_data_dir <-
file.path(tools::R_user_dir("gpttools", which = "data"), "text")
scraped_text_file <-
glue::glue("{scraped_data_dir}/{local_domain}.parquet")
if (file.exists(scraped_text_file) && rlang::is_false(overwrite)) {
cli::cli_abort(
c(
"!" = "Scraped data already exists for this domain.",
"i" = "Use {.code crawl(<url>, overwrite = TRUE)} to overwrite."
)
)
}
cli_rule("Crawling {.url {url}}")
cli_inform(c(
"i" = "This may take a while.",
"i" = "Gathering links to scrape"
))
links <-
recursive_hyperlinks(local_domain, url, aggressive = aggressive) |>
unique()
cli_inform(c("i" = "Scraping validated links"))
scraped_data <-
purrr::map(links, \(x) {
if (identical(check_url(x), 200L)) {
tibble::tibble(
link = x,
text = paste(scrape_url(x), collapse = " "),
n_words = tokenizers::count_words(text)
)
} else {
cli::cli_inform(c(
"!" = "Skipped {url}",
"i" = "Status code: {status}"
))
}
}) |>
dplyr::bind_rows() |>
dplyr::distinct()
arrow::write_parquet(
scraped_data,
glue("text/{local_domain}.parquet")
)
}recursive_hyperlinks <- function(local_domain,
url,
checked_urls = NULL,
aggressive = FALSE) {
links <- url[!(url %in% checked_urls)]
if (length(links) < 1) {
return(checked_urls)
}
if (aggressive) {
domain_pattern <- glue("^https?://(?:.*\\.)?{local_domain}/?")
} else {
domain_pattern <- glue("^https?://{local_domain}/?")
}
checked_urls <- c(checked_urls, links)
cli::cli_inform(c("i" = "Total urls: {length(checked_urls)}"))
links_df <- furrr::future_map(links, get_hyperlinks) |>
dplyr::bind_rows() |>
dplyr::filter(!stringr::str_detect(link, "^\\.$|mailto:|^\\.\\.|\\#|^\\_$"))
new_links <-
purrr::pmap(as.list(links_df), \(parent, link) {
clean_link <- NULL
if (stringr::str_detect(link, paste0("^https?://", local_domain))) {
clean_link <- link
} else if (stringr::str_detect(link, "^/[^/]|^/+$|^\\./|^[[:alnum:]]") &&
!stringr::str_detect(link, "^https?://")) {
if (stringr::str_detect(link, "^\\./")) {
clean_link <- stringr::str_replace(link, "^\\./", "/")
} else if (stringr::str_detect(link, "^[[:alnum:]]")) {
clean_link <- glue::glue("/", link)
} else {
clean_link <- link
}
clean_link <- glue::glue("{parent}{clean_link}")
}
}) |>
unlist()
recursive_hyperlinks(local_domain, unique(new_links), checked_urls)
}get_hyperlinks <- function(url) {
rlang::check_installed("rvest")
status <- httr::GET(url) |> httr::status_code()
if (identical(status, 200L)) {
tibble::tibble(
parent = url,
link = rvest::read_html(url) |>
rvest::html_nodes("a[href]") |>
rvest::html_attr("href") |>
unique()
)
} else {
cli::cli_warn(c(
"!" = "URL not valid.",
"i" = "Tried to scrape {url}",
"i" = "Status code: {status}"
))
}
}scrape_url <- function(url) {
rlang::check_installed("rvest")
exclude_tags <- c("style", "script", "head", "meta", "link", "button")
text <- rvest::read_html(url) |>
rvest::html_nodes(xpath = paste("//body//*[not(self::",
paste(exclude_tags, collapse = " or self::"),
")]",
sep = ""
)) |>
rvest::html_text2() |>
remove_new_lines()
if ("You need to enable JavaScript to run this app." %in% text) {
cli::cli_warn("Unable to parse page {url}. JavaScript is required.")
NULL
} else {
text
}
}crawl() that takes in a single argument, url, which is a character string of the URL to be scraped. The function scrapes all hyperlinks within the same domain. The scraped text is processed into a tibble format and saved as a parquet file using the {arrow} package into a directory called “text” with a filename that includes the local domain extracted earlier.
The function begins by extracting the local domain of the input URL using the urltools::url_parse() function, which returns a parsed URL object, and then extracting the domain component of the object.
The function then calls another function called recursive_hyperlinks() to recursively extract all hyperlinks within the url and validates the links in the process by only keeping urls that return a status code of 200 (i.e., the webpage is accessible).
The function then loops through each link and scrapes the text from each webpage and creates a tibble with three columns: link, text, and n_words. The link column contains the URL, the text column contains the scraped text, and the n_words column contains the number of words in the scraped text.
Generating Embeddings
After scraping the text data from the R4DS book, the next step is to generate embeddings for each chunk of text. This can be done using the create_index() function provided by the {gpttools} package. create_index() takes in a single argument, domain, which should be a character string indicating the domain of the scraped data.
Here’s the code to generate embeddings for the R4DS text data:
# Specify the domain of the scraped data
domain <- "r4ds.hadley.nz"
# Create embeddings for each chunk of text in the scraped data
index <- create_index(domain)create_index() function prepares the scraped data for indexing using the prepare_scraped_files() function, which splits the text into chunks and calculates the number of tokens in each chunk. It then calls add_embeddings() to generate embeddings for each chunk of text using the OpenAI API. A resulting tibble with embeddings is stored as a feather file using, again using the {arrow} package.
Here’s the code for the create_index() function and helper functions:
create_index <- function(domain, overwrite = FALSE) {
index_dir <-
file.path(tools::R_user_dir("gpttools", which = "data"), "index")
index_file <-
glue::glue("{index_dir}/{domain}.parquet")
if (file.exists(index_file) && rlang::is_false(overwrite)) {
cli::cli_abort(
c(
"!" = "Index already exists for this domain.",
"i" = "Use {.code overwrite = TRUE} to overwrite index."
)
)
}
cli::cli_inform(c(
"!" = "You are about to create embeddings for {domain}.",
"i" = "This will use many tokens. Only proceed if you understand the cost.",
"i" = "Read more about embeddings at {.url
https://platform.openai.com/docs/guides/embeddings}."
))
ask_user <- usethis::ui_yeah(
"Would you like to continue with creating embeddings?"
)
if (rlang::is_true(ask_user)) {
index <-
prepare_scraped_files(domain = domain) |>
add_embeddings()
if (rlang::is_false(dir.exists(index_dir))) {
dir.create(index_dir, recursive = TRUE)
}
arrow::write_parquet(
x = index,
sink = index_file
)
} else {
cli_inform("No index was created for {domain}")
}
}prepare_scraped_files <- function(domain) {
scraped_dir <- tools::R_user_dir("gpttools", which = "data")
arrow::read_parquet(glue("{scraped_dir}/text/{domain}.parquet")) |>
dplyr::mutate(
chunks = purrr::map(text, \(x) {
chunk_with_overlap(x,
chunk_size = 500,
overlap_size = 50,
doc_id = domain,
lowercase = FALSE,
strip_punct = FALSE,
strip_numeric = FALSE,
stopwords = NULL
)
})
) |>
tidyr::unnest(chunks) |>
tidyr::unnest(chunks) |>
dplyr::rename(original_text = text) |>
dplyr::mutate(n_tokens = tokenizers::count_characters(chunks) %/% 4)
}add_embeddings <- function(index) {
index |>
dplyr::mutate(
embeddings = purrr::map(
.x = chunks,
.f = create_openai_embedding,
.progress = "Create Embeddings"
)
) |>
tidyr::unnest(embeddings)
}create_openai_embedding <-
function(input_text,
model = "text-embedding-ada-002",
openai_api_key = Sys.getenv("OPENAI_API_KEY")) {
body <- list(
model = model,
input = input_text
)
embedding <- query_openai_api(body, openai_api_key, task = "embeddings")
embedding$usage$total_tokens
tibble::tibble(
usage = embedding$usage$total_tokens,
embedding = embedding$data$embedding
)
}The resulting tibble contains the following columns:
link: URL of the webpageoriginal_text: scraped textn_words: number of words in the scraped textchunks: text split into chunksusage: number of tokens used for creating the embeddingembeddings: embeddings for each chunk of text
Querying with Embeddings
After generating embeddings for each chunk of text, the next step is to query the embeddings to find similar chunks of text. This can be done using the query_index() function provided by the {gpttools} package. This is a bit of a complicated function, so it’s worth taking a look at the code to see how it works.
query_index <- function(index, query, history, task = "Context Only", k = 4) {
arg_match(task, c("Context Only", "Permissive Chat"))
query_embedding <- create_openai_embedding(input_text = query) |>
dplyr::pull(embedding) |>
unlist()
full_context <- get_top_matches(index, query_embedding, k = k)
context <-
full_context |>
dplyr::pull(chunks) |>
paste(collapse = "\n\n")
instructions <-
switch(task,
"Context Only" =
list(
list(
role = "system",
content =
glue(
"You are a helpful chat bot that answers questions based on the
context provided by the user. If the user does not provide
context, say \"I am not able to answer that question. Maybe
try rephrasing your question in a different way.\"\n\n
Context: {context}"
)
),
list(
role = "user",
content = glue("{query}")
)
),
"Permissive Chat" =
list(
list(
role = "system",
content =
glue(
"You are a helpful chat bot that answers questions based on the
context provided by the user. If the user does not provide
context, say \"I am not able to answer that question with the
context you gave me, but here is my best answer. Maybe
try rephrasing your question in a different way.\"\n\n
Context: {context}"
)
),
list(
role = "user",
content = glue("{query}")
)
)
)
cli_inform("Embedding...")
history <-
purrr::map(history, \(x) if (x$role == "system") NULL else x) |>
purrr::compact()
prompt <- c(history, instructions)
answer <- gptstudio::openai_create_chat_completion(prompt)
list(prompt, full_context, answer)
}get_top_matches <- function(index, query_embedding, k = 5) {
index |>
dplyr::mutate(similarity = purrr::map_dbl(embedding, \(x) {
lsa::cosine(query_embedding, unlist(x))
})) |>
dplyr::arrange(dplyr::desc(similarity)) |>
head(k)
}openai_create_chat_completion <-
function(prompt = "<|endoftext|>",
model = "gpt-3.5-turbo",
openai_api_key = Sys.getenv("OPENAI_API_KEY"),
task = "chat/completions") {
assert_that(
is.string(model),
is.string(openai_api_key)
)
if (is.string(prompt)) {
prompt <- list(
list(
role = "user",
content = prompt
)
)
}
body <- list(
model = model,
messages = prompt
)
query_openai_api(body, openai_api_key, task = task)
}query_openai_api <- function(body, openai_api_key, task) {
arg_match(task, c("completions", "chat/completions", "edits", "embeddings"))
base_url <- glue("https://api.openai.com/v1/{task}")
headers <- c(
"Authorization" = glue("Bearer {openai_api_key}"),
"Content-Type" = "application/json"
)
response <-
httr::RETRY("POST",
url = base_url,
httr::add_headers(headers), body = body,
encode = "json",
quiet = TRUE
)
parsed <- response %>%
httr::content(as = "text", encoding = "UTF-8") %>%
jsonlite::fromJSON(flatten = TRUE)
if (httr::http_error(response)) {
cli_alert_warning(c(
"x" = glue("OpenAI API request failed [{httr::status_code(response)}]."),
"i" = glue("Error message: {parsed$error$message}")
))
}
parsed
}The query_index() function takes in four arguments: index, query, task, and k.
index: The index containing the embeddings to be queried.query: The query string to search for similar chunks of text.task: The type of task to perform based on the context of the query. {gpttools} provides a few pre-defined tasks, such as “conservative q&a” or “extract key libraries and tools”, which can be passed to this argument. These queries were taken from a repo created by OpenAI and Pinecone, which can be found here.k: The number of similar chunks of text to return.
The function generates an embedding for the query string with create_openai_embedding(). It then uses the get_top_matches() to find the most similar chunks of text in the index using cosine similarity returning the top k matches.
The next step is to formats the instructions based on the task argument as well as the query and context. For example, if the task is “conservative q&a”, the function will return a string asking the model to answer a question based on the context of the returned chunks of text. If the task is “extract key libraries and tools”, the function will return a string listing the libraries and tools present in the returned chunks of text.
The prompt is passed to OpenAI’s GPT-3.5 gpt-3.5-turbo (i.e., ChatGPT) to generate a response based on the formatted output. The response is returned as a list containing the instructions, the top k matches, and the response from ChatGPT.
Analysis of Embeddings with tidymodels and UMAP
The next step is to perform dimension reduction with UMAP as naive exploration of the embeddings. The embeddings column in the index contains a list of 1536-dimensional vectors. We can use the recipes package to normalize the vectors and then use the step_umap() function to reduce the dimensionality to 2. We can then plot the results.
library(recipes)
library(ggplot2)
library(purrr)
library(tidyr)
library(embed)
index_wide <-
index |>
mutate(embedding = map(embedding, \(x) as.data.frame(t(x)))) |>
unnest(embedding)
set.seed(123)
umap_spec <- recipe(~ ., data = index_wide) |>
step_normalize(starts_with("V")) |>
step_umap(starts_with("V"), num_comp = 2)
umap_estimates <- prep(umap_spec, training = index_wide)
umap_data <- bake(umap_estimates, new_data = NULL)
umap_data |>
ggplot() +
geom_point(aes(x = UMAP1, y = UMAP2, color = link),
alpha = 0.5, size = 2, show.legend = FALSE) +
labs(title = "Dimensionality Reduction with UMAP",
subtitle = "UMAP of 1536-dimensional vectors | Colored by Source Text") +
theme_minimal()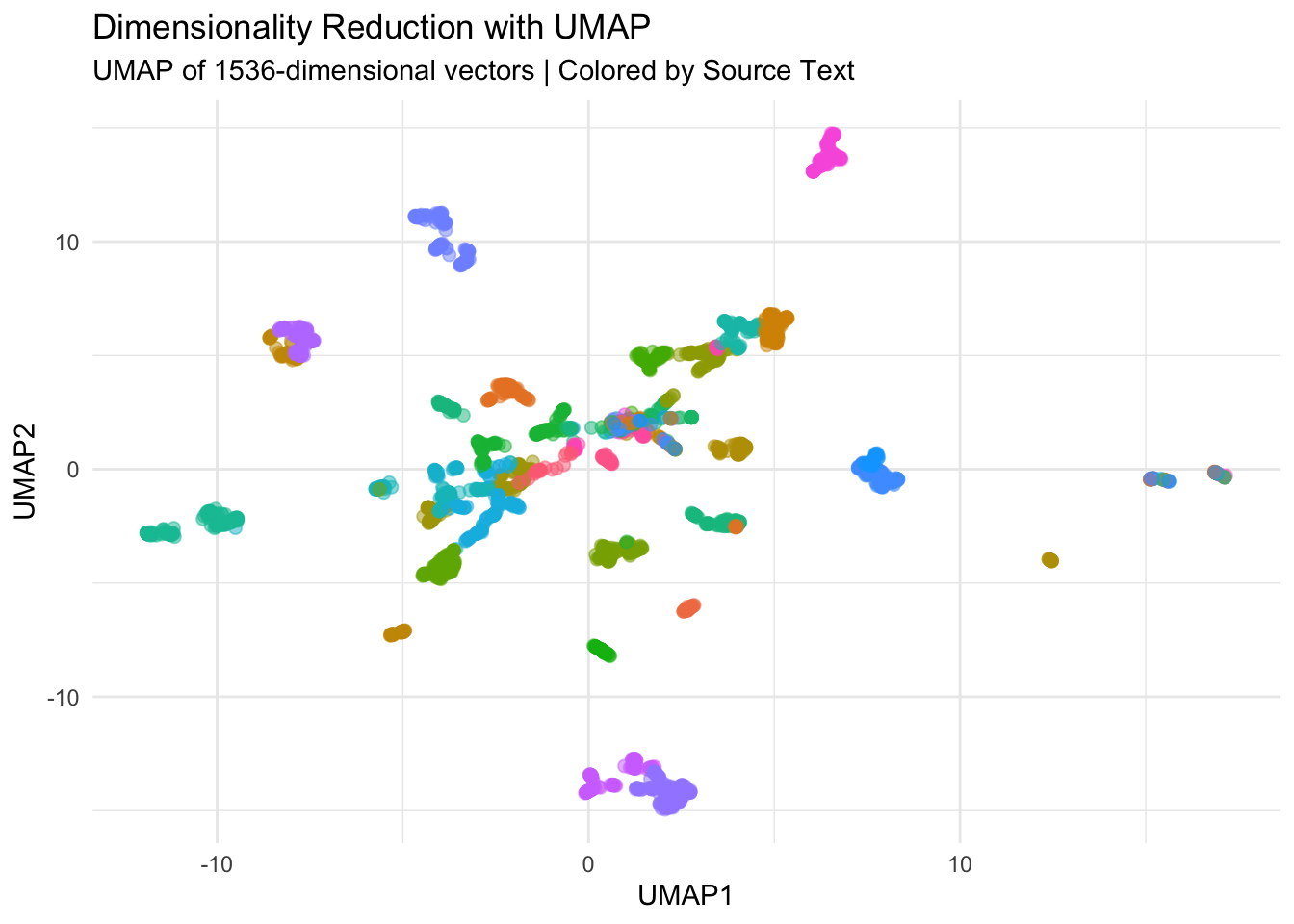
Retreiver Plugin as a Shiny App
To use the index, {gpttools} now as a shiny app that you can run as a plugin in RStudio. To use it, open the command palette (Cmd/Ctrl + Shift + P) and type “gpttools”. You can then select the “gpttools: ChatGPT with Retrieval” option. This will open a shiny app in your viewer pane. If you have multiple indices, you can select which one to use in the dropdown menu. You can also specify if you want answers that only use the context of index (“Context Only”) or if you want answers that use the context of the index and full ChatGPT model (“Permissive Chat”). “Context Only” answers are much less likely to “hallucinate.” “Permissive Chat” answers are more likely to hallucinate, but they are also more likely choice if the index lacks relevant information.
Photo Credit
Thumbnail Photo by Richard Burlton on Unsplash